Prokaryotes have evolved a diverse arsenal of immune systems that provide protection against the constant threat of invading nucleic acids such as viruses and plasmids. In the last decade, numerous different prokaryotic immune systems (e.g. CRISPR-Cas and prokaryotic Argonaute proteins) have been characterized. An important aspect of the functionality of both CRISPR-Cas and Argonaute systems is that the effector proteins of these systems can be (re)programmed with small nucleic-acid guides. Such guides allow for sequence-specific recognition of invading DNA.
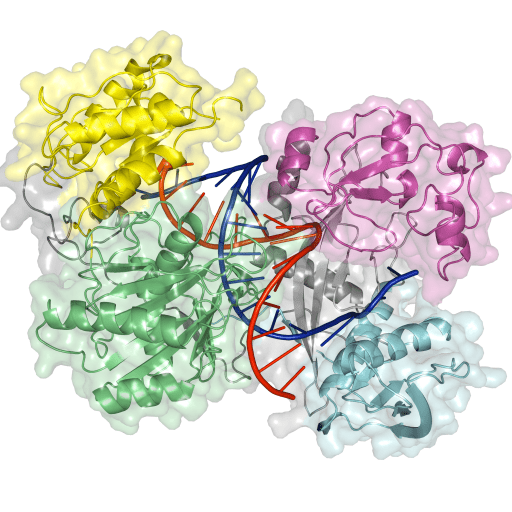
DNA guides to bind and cleave DNA targets
These prokaryotic immune systems are not only interesting from fundamental biology and evolutionary biology perspectives, but their utilization as programmable genome editing tools or nucleic acid detection tool caused a revolution in basic biology research. Furthermore, these tools have a great potential for diagnostic purposes and to treat genetic diseases in the future.
Although certain prokaryotic immune systems are well studied, there is tremendous diversity, and most systems remain poorly characterized.
We are interested in the diversity of prokaryotic immune systems from a fundamental perspective, but also look into the possibilities of repurposing them as molecular tools.
Research questions
In the Swarts lab, we focus on the following research questions:
- How do different prokaryotic immune systems protect their host?
- How are the macromolecular architecture and functionality of these systems connected?
- How do prokaryotic immune systems distinguish between invaders and self?
- Can prokaryotic immune systems be repurposed as molecular tools?
Techniques
To address our research questions, we employ research techniques from various fields ranging from bacterial genetics and subsequent in vivo functional characterization, to in vitro protein biochemistry. In addition, we use X-ray crystallography to obtain insights in structure and protein-nucleic acid interactions to reveal mechanisms underlying the functionality of the proteins we study. Combined, this allows us to chart in detail the function, structure, and biochemical mechanisms of prokaryotic immune systems.